
As you can see from our publications, we love to research and collaborate on a variety of projects related to protein folding, chaperones and neurodegeneration. We actively look for ways to share our expertise in a variety of biochemical and cellular techniques to complement the awesome skills of other researchers so that we can answer very exciting research questions together.
2024 Cell Parra Bravo C, Giani AM, Madero-Perez J, Zhao Z, Wan Y, Samelson AJ, Wong MY, Evangelisti A, Cordes E, Fan L, Ye P, Zhu D, Pozner T, Mercedes M, Patel T*, Yarahmady A*, Carling GK, Sterky FH, Lee VMY, Lee EB, DeTure M, Dickson DW, Sharma M, Mok SA, Luo W, Zhao M, Kampmann M, Gong S, Gan L. Human iPSC 4R tauopathy model uncovers modifiers of tau propagation
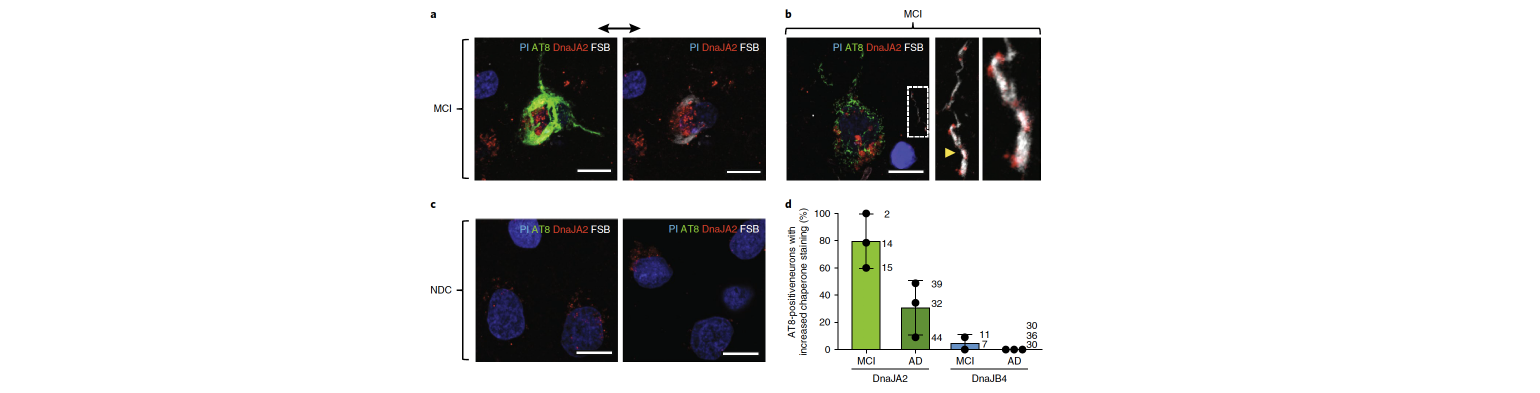
2023 ACS Chemical Neuroscience Sun KT*, Patel T*, Kang S-G*, Yarahmady A*, Julien O, Heras J, Mok SA. Disease associated mutations in tau encode for changes in aggregate structure conformation
2023 Scientific Reports Paul PS, Patel T*, Cho J-Y, Yarahmady A*, Khalili A, Semencheko V, Wille H, Kulka M, Mok SA, Kar S. Native PLGA nanoparticles attenuate Aβ-seed induced tau aggregation under in vitro conditions: potential implication in Alzheimer's disease pathology.
2023 Nature Neuroscience Udeochu J, Amin S, Huang Y, Fan L, Torres E, Liu B, McGurran H, Coronas-Samano G, Kauwe G, Mousa GA, Wong MY, Ye P, Nagiri RK, Lo I, Holtzman J, Corona C, Yarahmady A*, Gill M, Raju R, Mok SA, Gong S, Luo W, Tracy TE, Ratan R , Tsai L-H, Sinha S, Gan L. Tau activation of microglial cGAS–IFN reduces MEF2C-mediated cognitive resilience.
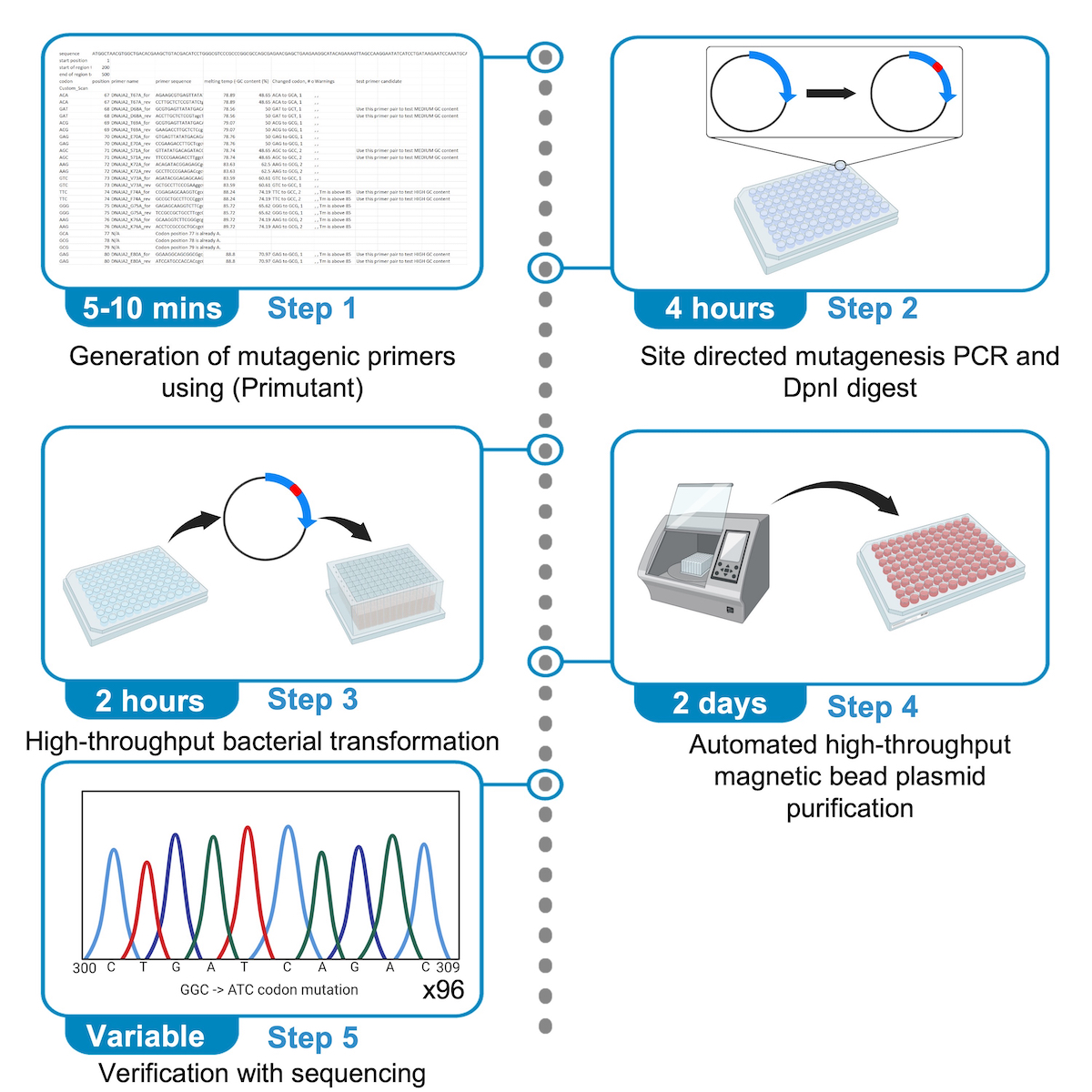
2022 STAR Protocols Sun KT*, Patel TS*, Kim J*, Tang HSH, Eskandari-Sedighi G, Sureshkumar H*, Schieve D, Mok SA. Streamlined high-throughput cloning protocol to generate arrayed mutant libraries
2022 Cell Tracy TE, Madero-Pérez J, Swaney DL, Chang TS, Moritz M, Konrad C, Ward ME, Stevenson E, Hüttenhain R, Kauwe G, Mercedes M, Sweetland-Martin L, Chen X, Mok SA, Wong MY, Telpoukhovskaia M, Min SW, Wang C, Sohn PD, Martin J, Zhou Y, Luo W, Trojanowski JQ, Lee VMY, Gong S, Manfredi G, Coppola G, Krogan NJ, Geschwind DH, Gan L. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration
2022 Nature Communications Wang C, Fan L, Khawaja RR, Liu B, Zhan L, Kodama L, Chin M, Li Y, Le D, Zhou Y, Condello C, Grinberg LT, Seeley WW, Miller BL, Mok SA, Gestwicki JE, Cuervo AM, Luo W, Gan L. Microglial NF-κB drives tau spreading and toxicity in a mouse model of tauopathy
2021 BMC Biology Kang SG, Han ZZ, Daude N, McNamara E*, Wohlgemuth S, Molina-Porcel L, Safar JG, Mok SA and Westaway D. Pathologic tau conformer ensembles induce dynamic, liquid-liquid phase separation events at the nuclear envelope
2021 Frontiers in Genetics Budrass L*, Fahlman R, Mok SA. Deciphering Network Crosstalk: The Current Status and Potential of miRNA Regulatory Networks on the HSP40 Molecular Chaperone Network
2020 Molecular Neurodegeneration Chen X, Li Y, Wang C, Tang Y, Mok SA, Tsai RM, Rojas JC, Karydas A, Miller BL, Boxer AL, Gestwicki JE, Arkin M, Cuervo AM, Gan L. Promoting tau secretion and propagation by hyperactive p300/CBP via autophagy-lysosomal pathway in tauopathy
2019 Journal of Biological Chemistry Chen JJ, Nathaniel DL, Raghavan P, Nelson M, Tian R, Tse E, Hong JY, See SK, Mok SA, Hein MY, Southworth DR, Grinberg LT, Gestwicki JE, Leonetti MD, Kampmann M. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation
2019 Neuron Sohn PD, Huang CT, Yan R, Fan L, Tracy TE, Camargo CM, Montgomery KM, Arhar T, Mok SA, Freilich R, Baik J, He M, Gong S, Roberson ED, Karch CM, Gestwicki JE, Xu K, Kosik KS, Gan L. Pathogenic Tau Impairs Axon Initial Segment Plasticity and Excitability Homeostasis
2019 Nature Communications Freilich R., Betegon M., Tse E., Mok S.A., Julien O., Agard D.A., Southworth D.R., Takeuchi K., Gestwicki J.E. Competing protein-protein interactions regulate binding of Hsp27 to its client protein tau
2018 Nature Structure & Molecular Biology Mok S.A., Condello C., Freilich R., Gillies, A., Arhar T., Oroz J., Kadavath H., Julien O., Assimon V.A., Rauch J.N., Dunyak B.M., Lee J., Tsai F.T.F., Wilson M.R., Zweckstetter M., Dickey C.A., Gestwicki J.E. Mapping Interactions with the Chaperone Network Reveals Proteostasis Factors that Protect Against Tau Aggregation
2018 Journal of Neuroscience Min S-W, Sohn P.D., Li T., Devidze N., Johnson J.R., Krogan N.J., Masliah E., Mok S.A., Gestwicki J.E. Gan L. SIRT1 Deacetylates Tau and Reduces Pathogenic Tau Spread in a Mouse Model of Tauopathy
2018 Journal of Huntington's Disease Chen M.Z., Mok S.A., Muchowski P.J., Hatters D.M. N-terminal fragments of Huntingtin longer than residue 170 form visible aggregates independently to polyglutamine expansion
2017 Journal of Molecular Biology Rauch, J.N., Tse E., Freilich, R., Mok S.A., Makley, L.N., Southworth D.R., Gestwicki, J.E. BAG3 is a modular scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins
2017 Cold Spring Harbor Perspectives in Medicine Young, Z. T., Mok, S.A., Gestwicki, J. E.Therapeutic strategies for restoring tau homeostasis
2015 Nature Medicine Min S-W., Chen X., Tracy T.E., Li Y., Zhou Y., Wang C., Shirakawa K., Minami S.S., Defensor E., Mok S.A., Dongmin P., Sohn P.D., Cong X., Ellerby L., Gibson B.W., Johnson J., Krogan N., Shamloo M., Gestwicki J.E., Masliah E., Verdin E., Gan L. Critical Role of Acetylation in Tau-Mediated Neurodegeneration and Cognitive Deficits
2014 Chemical Biology & Drug Design Walter G.M., Raveh A., Mok S.A., McQuade T.J., Arevang C.J., Schultz P.J., Smith M.C., Asare S., Cruz P.G., Wisen S., Matainaho T., Sherman D.H., Gestwicki J.E. High Throughput Screen of Natural Product Extracts in A Yeast Model of Polyglutamine Proteotoxicity
During my PhD work I focused on elucidating mechanisms by which Nerve Growth Factor (NGF) regulates the survival of neurons. I had a special interest in long distance signaling pathways that travel between the axon terminals and the nucleus. I was very priviledged to be able to train under the mentorship of Dr. Bob campenot who invented the famous Campenot culture system for neurons. This system allowed us to specifically apply treatments to the axons or cell bodies of neurons and easily analyze effects over time.
2013 Journal of Neuroscience Methods Mok, S.A., Lund K., LaPointe P., Campenot R.B. A HaloTag Method for Assessing the Axonal Transport of Membrane Proteins and Intracellular Proteins in Compartmented Cultures of Sympathetic Neurons
2009 Nature Protocols Campenot, R.B., Lund K., Mok S.A. Production of compartmented cultures of rat sympathetic neurons
2009 Cell Research Mok, S.A., Lund K., Campenot R.B. A retrograde apoptotic signal originating in NGF-deprived distal axons of rat sympathetic neurons in compartmented cultures
2007 Neuropharmacology Mok, S.A., Campenot R.B. A nerve growth factor-induced retrograde survival signal mediated by mechanisms downstream of TrkA
September 1, 2024
Joining our team this fall are Scott Dixon (graduate student), Ashlyn Benko (graduate student), Grace Cassidy-Ketchin (undergraduate) and Angela Lebrudo (undergraduate). See our people page for more info.
August 14, 2024
We had fun this summer with a full team in the lab.
We welcomed 4 undergraduate summer students (Shreya Ghosh, Alicia Guzman, Natasha Larson, Lisa Wang), as well as Brielle Huang (high school student and local Brain Bee winner).
Pictures from our Summer 2024 escape room outing:


March 14, 2024
We have been busy but it is about time we highlighted some of the exciting things that have happened in the last year:
Our team in collaboration with the Heras and Condello labs published a study on the development of our high-throughput tau protein purification methods and trypsin digest assay for identifying structure differences between tau aggregates. We were able to use our new techniques to show that tau missense mutations associated with neurodegeneration can alter the inherent aggregate structures formed in vitro. Our findings have potential implications for tauopathies caused by tau mutations and interpreting data from many of the experimental models we use to study tauopathies. You can check out the study at ACS Chemical Neuroscience or BioRxiv.
Our team (Mok, Condello, and Heras labs) was awarded a CIHR project grant to fund our work to generate new tau aggregate structures and characterize their effects in cell and animal models!

We have added several new members to our team who are contributing their skills and enthusiasm to our research program.

Our most recent lab photo. Left to Right: Sang-Gyun Kang, Kerry Sun, Varun Aggarwal, Mahalashmi Srinivasan, Ria Ratra, Jenna Lindner, Sue-Ann Mok
Pictures from Summer 2023 go karting with the lab:


The lab had a great time attending the PRinCE 2023 meeting in Montreal. What a great research community to be a part of!


November 28, 2022
With an idea that started in our homes at the beginning of the pandemic came our paper detailing how to make arrayed mutant libraries which is now published at STAR Protocols! We hope that the paper accomplishes our goal of giving any standard molecular biology lab the tools to make custom arrayed mutant libraries economically and rapidly. Check out our free primer design webapp page with links to the protocol paper!
July 22, 2022
Our entire team thanks the donors of the BrightFocus Foundation for recently supporting our study to look at the role of tau phosphorylation sites on tau aggregate structure and pathology. We are working with an incredible team that includes Carlo Condello (UCSF), HyunJun Yang (UCSF), and Jonathan Heras (University de La Rioja). Get ready for some exciting results!
Click here to read more about the incredible research funded by the BrightFocus Foundation.