PRIMUTANT was developed by a team of undergraduate and graduate students from the Department of Biochemistry at the University of Alberta under the supervision of Sue-Ann Mok and Dean Schieve. The site was initially launched in October 2020 and is continuously being updated and maintained.
You can access the program with our easy-to-use Webapp found on the homepage. You can also download all scripts from GitHub and modify the program for your own projects as you see fit.
Primer Design Parameters
The organism selection (mammalian/bacterial) reflects that in certain organisms, some codons have low levels of their respective tRNAs. To account for this, this program avoids using the these rare codons for the mutations. For codon usage, rare codons (less than 0.11 fraction of all codons for a given amino acid) based on available data from Genscript are excluded from being selected as substituting mutations in primers.
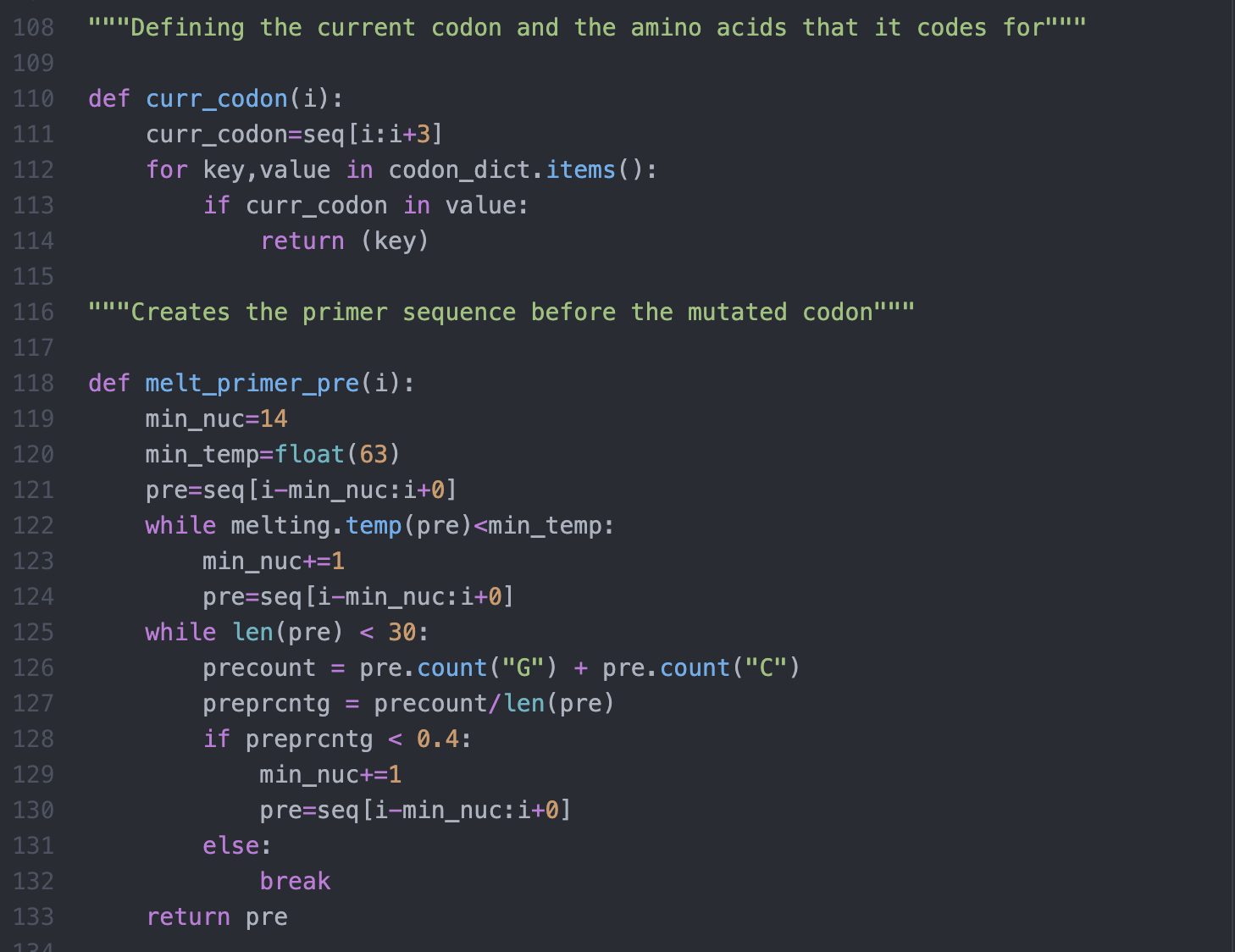
The primer design is handled by a variety of functions that takes Tm, length and GC content into account, and also checks for potential complications such as hairpins and homo/heterodimers. The current parameters that the program uses are:
Tm: Min: 63°C and warning given after 85°C.
Length: Minimum of 14 nucleotides before and after the codon of interest. Maximum addition of 30 nucleotides before and after the codon of interest.
GC content: Ideal GC content of 40% to 60%. Warnings are given if GC content is less that 40% or greater than 60%.
Nucleotide changes minimized.
Summary of Mutagenesis Options
Scanning site-saturation mutagenesis: This option allows for the substitution of each amino acid in the region of interest with every other possibility of amino acid. The program changes the codon so that it bears the least nucleotide changes to the original sequence.
Custom scanning mutagenesis: This option allows for the substitution of each amino acid in the region of interest with the amino acid of the user's choice. Similarly to scanning site-saturation mutagenesis, the program changes the codon so that it bears the least changes to the original sequence.
Phosphorylation site mutagenesis: This option has two categories: Phosphomimetic/Phosphorylation-deficient site changes. Phosphomimetic option changes serine and threonine residues to glutamate residues, as it bears the closest similarity to phospho-serine. Phosphorylation-deficient option changes all possible tyrosine, serine and threonine residues to alanine.